rmcmc is an R package for simulating Markov chains using
the Barker proposal to compute Markov chain Monte Carlo (MCMC)
estimates of expectations with respect to a target distribution on a
real-valued vector space. The Barker proposal, described in Livingstone and Zanella
(2022), is a gradient-based MCMC algorithm inspired by the Barker
accept-reject rule. It combines the robustness of simpler MCMC schemes,
such as random-walk Metropolis, with the efficiency of gradient-based
methods, such as the Metropolis adjusted Langevin algorithm.
The key function provided by the package is
sample_chain(), which allows sampling a Markov chain with a
specified target distribution as its stationary distribution. The chain
is sampled by generating proposals and accepting or rejecting them using
a Metropolis-Hasting acceptance rule. During an initial warm-up stage,
the parameters of the proposal distribution can be adapted, with
adapters available to both: tune the scale of the proposals by coercing
the average acceptance rate to a target value; tune the shape of the
proposals to match covariance estimates under the target distribution.
As well as the default Barker proposal, the package also provides
implementations of alternative proposal distributions, such as
(Gaussian) random walk and Langevin proposals. Optionally, if BridgeStan’s
R interface, available on GitHub, is
installed, then
BridgeStan can be used to specify the target distribution to sample
from.
The latest published release version of rmcmc on CRAN
can be installed using
install.packages("rmcmc")Alternatively, the current development version of rmcmc
can be installed using
# install.packages("devtools")
devtools::install_github("UCL/rmcmc")The snippet below shows a basic example of using the package to generate samples from a normal target distribution with random scales. Adapters are used to tune the proposal scale to achieve a target average acceptance probability, and to tune the proposal shape with per-dimension scale factors based on online estimates of the target distribution variances.
library(rmcmc)
set.seed(876287L)
dimension <- 3
scales <- exp(rnorm(dimension))
target_distribution <- list(
log_density = function(x) -sum((x / scales)^2) / 2,
gradient_log_density = function(x) -x / scales^2
)
proposal <- barker_proposal()
results <- sample_chain(
target_distribution = target_distribution,
initial_state = rnorm(dimension),
n_warm_up_iteration = 10000,
n_main_iteration = 10000,
proposal = proposal,
adapters = list(scale_adapter(), shape_adapter("variance"))
)
mean_accept_prob <- mean(results$statistics[, "accept_prob"])
adapted_shape <- proposal$parameters()$shape
cat(
sprintf("Average acceptance probability is %.2f", mean_accept_prob),
sprintf("True target scales: %s", toString(scales)),
sprintf("Adapter scale est.: %s", toString(adapted_shape)),
sep = "\n"
)
#> Average acceptance probability is 0.58
#> True target scales: 1.50538046096953, 1.37774732725824, 0.277038897322645
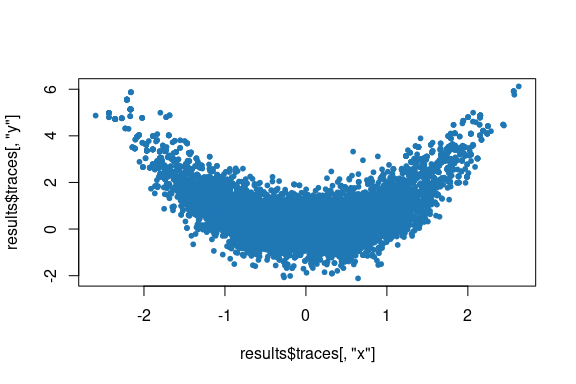
#> Adapter scale est.: 1.5328097767097, 1.42342707172926, 0.280359693392091As a second example, the snippet below demonstrates sampling from a
two-dimensional banana shaped distribution based on the Rosenbrock
function and plotting the generated chain samples. Here we use the
default values of the proposal and adapters
arguments to sample_chain(), corresponding respectively to
the Barker proposal, and adapters for tuning the proposal scale to
coerce the average acceptance rate using a dual-averaging algorithm, and
for tuning the proposal shape based on an estimate of the target
distribution covariance matrix. The target_distribution
argument to sample_chain() is passed a formula specifying
the log density of the target distribution, which is passed to
target_distribution_from_log_density_formula() to construct
necessary functions, using stats::deriv() to symbolically
compute derivatives.
library(rmcmc)
set.seed(651239L)
results <- sample_chain(
target_distribution = ~ (-(x^2 + y^2) / 8 - (x^2 - y)^2 - (x - 1)^2 / 100),
initial_state = rnorm(2),
n_warm_up_iteration = 10000,
n_main_iteration = 10000
)
plot(results$traces[, "x"], results$traces[, "y"], col = "#1f77b4", pch = 20)