Plan · Analyze · Explore

Install from CRAN:
install.packages("multitool")You can install the development version of multitool
from GitHub with:
# install.packages("devtools")
devtools::install_github("ethan-young/multitool")The goal of multitool is to provide a set of tools for
designing and running multiverse-style analyses. I designed it to help
users create an incremental workflow for slowly building up, keeping
track of, and unpacking multiverse analyses and results.
For those unfamiliar with multiverse analysis, here is a short primer:
I designed multitool to do multiverse analysis but its
really just a tool for exploration.
In any new field, area, or project, there is a lot of uncertainty
about which data analysis decisions to make. Clear research questions
and criteria help reduce uncertainty about how to answer them but they
never fully reduce them. multitool helps organize and
systematically explore different options. That’s really it.
I designed multitool to help users take a single use
case (e.g., a single analysis pipeline) and expand it into a workflow to
include alternative versions of the same analysis.
For example, imagine you would like to take some data, remove
outliers, transform variables, run a linear model, do a post-hoc
analysis, and plot the results. multitool can take theses
tasks and transform them into a blueprint, which provides instructions
for running your analysis pipeline.
The functions were designed to play nice with the tidyverse and require
using the base R pipe |>. This makes it easy to quickly
convert a single analysis into a multiverse analysis.
My vision of a multitool workflow contains five
steps:
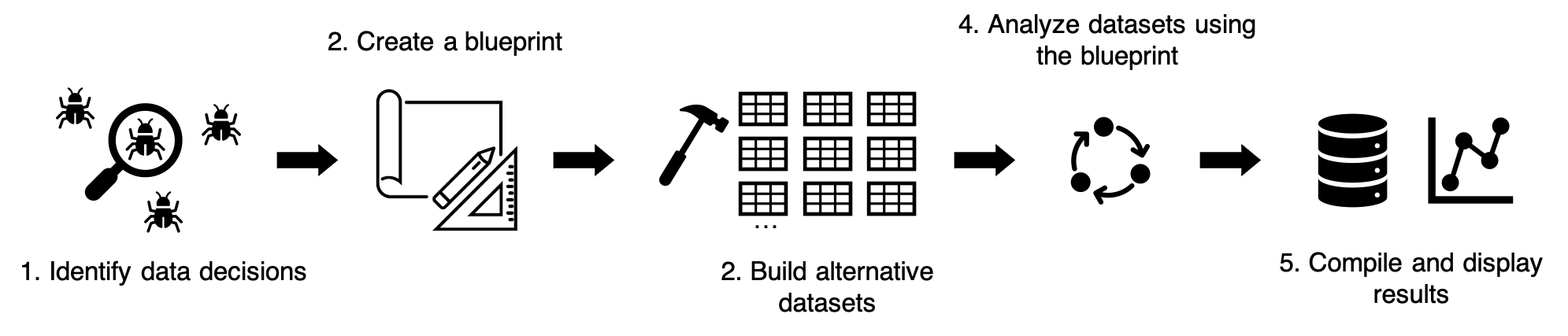
multitool cannot make decisions for you but – once you
know your set of data decisions – it can help you create and organize
them into the workflow above.
A defining feature of multitool is that it saves your
code. This allows the user to grab the code that produces a
result and inspect it for accuracy, errors, or simply for peace of
mind. By quickly grabbing code, the user can iterate between creating
their blueprint and checking that the code works as intended.
multitool allows the user to model data however they’d
like. The user is responsible for loading the relevant modeling
packages. Regardless of your model choice, multitool will
capture your code and build a blueprint with alternative analysis
pipelines.
Finally, multiverse analyses were originally intended to look at how model parameters shift as a function of arbitrary data decisions. However, any computation might change depending on how you slice and dice the data. For this reason, I also built functions for computing descriptive, correlation, and reliability analysis alongside a particular modelling pipeline.
# load packages
library(tidyverse)
library(multitool)
# create some data
the_data <-
data.frame(
id = 1:100,
iv1 = rnorm(100),
iv2 = rnorm(100),
iv3 = rnorm(100),
mod = rnorm(100),
dv1 = rnorm(100),
dv2 = rnorm(100),
include1 = rbinom(100, size = 1, prob = .1),
include2 = sample(1:3, size = 100, replace = TRUE),
include3 = rnorm(100)
)
# create a pipeline blueprint
full_pipeline <-
the_data |>
add_filters(include1 == 0, include2 != 3, include3 > -2.5) |>
add_variables(var_group = "ivs", iv1, iv2, iv3) |>
add_variables(var_group = "dvs", dv1, dv2) |>
add_model("linear model", lm({dvs} ~ {ivs} * mod))
full_pipeline
#> # A tibble: 12 × 4
#> type group code additional_args
#> <chr> <chr> <chr> <lgl>
#> 1 filters include1 include1 == 0 NA
#> 2 filters include1 include1 %in% unique(include1) NA
#> 3 filters include2 include2 != 3 NA
#> 4 filters include2 include2 %in% unique(include2) NA
#> 5 filters include3 include3 > -2.5 NA
#> 6 filters include3 include3 %in% unique(include3) NA
#> 7 variables ivs iv1 NA
#> 8 variables ivs iv2 NA
#> 9 variables ivs iv3 NA
#> 10 variables dvs dv1 NA
#> 11 variables dvs dv2 NA
#> 12 models linear model lm({dvs} ~ {ivs} * mod) NA
# Expand your blueprint into a grid
expanded_pipeline <- expand_decisions(full_pipeline)
expanded_pipeline
#> # A tibble: 48 × 4
#> decision variables filters models
#> <chr> <list> <list> <list>
#> 1 1 <tibble [1 × 2]> <tibble [1 × 3]> <tibble [1 × 3]>
#> 2 2 <tibble [1 × 2]> <tibble [1 × 3]> <tibble [1 × 3]>
#> 3 3 <tibble [1 × 2]> <tibble [1 × 3]> <tibble [1 × 3]>
#> 4 4 <tibble [1 × 2]> <tibble [1 × 3]> <tibble [1 × 3]>
#> 5 5 <tibble [1 × 2]> <tibble [1 × 3]> <tibble [1 × 3]>
#> 6 6 <tibble [1 × 2]> <tibble [1 × 3]> <tibble [1 × 3]>
#> 7 7 <tibble [1 × 2]> <tibble [1 × 3]> <tibble [1 × 3]>
#> 8 8 <tibble [1 × 2]> <tibble [1 × 3]> <tibble [1 × 3]>
#> 9 9 <tibble [1 × 2]> <tibble [1 × 3]> <tibble [1 × 3]>
#> 10 10 <tibble [1 × 2]> <tibble [1 × 3]> <tibble [1 × 3]>
#> # ℹ 38 more rows
# Run the blueprint
multiverse_results <- run_multiverse(expanded_pipeline)
multiverse_results
#> # A tibble: 48 × 4
#> decision specifications model_fitted pipeline_code
#> <chr> <list> <list> <list>
#> 1 1 <tibble [1 × 3]> <tibble [1 × 5]> <tibble [1 × 2]>
#> 2 2 <tibble [1 × 3]> <tibble [1 × 5]> <tibble [1 × 2]>
#> 3 3 <tibble [1 × 3]> <tibble [1 × 5]> <tibble [1 × 2]>
#> 4 4 <tibble [1 × 3]> <tibble [1 × 5]> <tibble [1 × 2]>
#> 5 5 <tibble [1 × 3]> <tibble [1 × 5]> <tibble [1 × 2]>
#> 6 6 <tibble [1 × 3]> <tibble [1 × 5]> <tibble [1 × 2]>
#> 7 7 <tibble [1 × 3]> <tibble [1 × 5]> <tibble [1 × 2]>
#> 8 8 <tibble [1 × 3]> <tibble [1 × 5]> <tibble [1 × 2]>
#> 9 9 <tibble [1 × 3]> <tibble [1 × 5]> <tibble [1 × 2]>
#> 10 10 <tibble [1 × 3]> <tibble [1 × 5]> <tibble [1 × 2]>
#> # ℹ 38 more rows
# Unpack model coefficients
multiverse_results |>
reveal_model_parameters()
#> # A tibble: 192 × 20
#> decision specifications model_function parameter unstd_coef se unstd_ci
#> <chr> <list> <chr> <chr> <dbl> <dbl> <dbl>
#> 1 1 <tibble [1 × 3]> lm (Intercep… 0.145 0.119 0.95
#> 2 1 <tibble [1 × 3]> lm iv1 0.117 0.105 0.95
#> 3 1 <tibble [1 × 3]> lm mod -0.0717 0.122 0.95
#> 4 1 <tibble [1 × 3]> lm iv1:mod 0.0833 0.126 0.95
#> 5 2 <tibble [1 × 3]> lm (Intercep… 0.0996 0.114 0.95
#> 6 2 <tibble [1 × 3]> lm iv1 0.218 0.101 0.95
#> 7 2 <tibble [1 × 3]> lm mod 0.213 0.117 0.95
#> 8 2 <tibble [1 × 3]> lm iv1:mod -0.00526 0.121 0.95
#> 9 3 <tibble [1 × 3]> lm (Intercep… 0.121 0.115 0.95
#> 10 3 <tibble [1 × 3]> lm iv2 0.0323 0.117 0.95
#> # ℹ 182 more rows
#> # ℹ 13 more variables: unstd_ci_low <dbl>, unstd_ci_high <dbl>, t <dbl>,
#> # df_error <int>, p <dbl>, std_coef <dbl>, std_ci <dbl>, std_ci_low <dbl>,
#> # std_ci_high <dbl>, model_performance <list>, model_warnings <list>,
#> # model_messages <list>, pipeline_code <list>
# Unpack model fit statistics
multiverse_results |>
reveal_model_performance()
#> # A tibble: 48 × 14
#> decision specifications model_function model_parameters aic aicc bic
#> <chr> <list> <chr> <list> <dbl> <dbl> <dbl>
#> 1 1 <tibble [1 × 3]> lm <prmtrs_m> 158. 159. 168.
#> 2 2 <tibble [1 × 3]> lm <prmtrs_m> 153. 154. 164.
#> 3 3 <tibble [1 × 3]> lm <prmtrs_m> 157. 158. 167.
#> 4 4 <tibble [1 × 3]> lm <prmtrs_m> 158. 159. 168.
#> 5 5 <tibble [1 × 3]> lm <prmtrs_m> 159. 160. 169.
#> 6 6 <tibble [1 × 3]> lm <prmtrs_m> 153. 154. 163.
#> 7 7 <tibble [1 × 3]> lm <prmtrs_m> 158. 159. 168.
#> 8 8 <tibble [1 × 3]> lm <prmtrs_m> 153. 154. 164.
#> 9 9 <tibble [1 × 3]> lm <prmtrs_m> 157. 158. 167.
#> 10 10 <tibble [1 × 3]> lm <prmtrs_m> 158. 159. 168.
#> # ℹ 38 more rows
#> # ℹ 7 more variables: r2 <dbl>, r2_adjusted <dbl>, rmse <dbl>, sigma <dbl>,
#> # model_warnings <list>, model_messages <list>, pipeline_code <list>
# Summarize model coefficients
multiverse_results |>
reveal_model_parameters() |>
group_by(parameter) |>
condense(unstd_coef, list(mean = mean, median = median, sd = sd))
#> # A tibble: 8 × 5
#> parameter unstd_coef_mean unstd_coef_median unstd_coef_sd unstd_coef_list
#> <chr> <dbl> <dbl> <dbl> <list>
#> 1 (Intercept) 0.105 0.0999 0.0365 <dbl [48]>
#> 2 iv1 0.160 0.155 0.0740 <dbl [16]>
#> 3 iv1:mod 0.0159 -0.00955 0.0622 <dbl [16]>
#> 4 iv2 0.00641 0.00608 0.0659 <dbl [16]>
#> 5 iv2:mod 0.0789 0.0776 0.0750 <dbl [16]>
#> 6 iv3 -0.0205 -0.0321 0.103 <dbl [16]>
#> 7 iv3:mod 0.0812 0.0827 0.175 <dbl [16]>
#> 8 mod 0.0802 0.0808 0.108 <dbl [48]>
# Summarize fit statistics
multiverse_results |>
reveal_model_performance() |>
condense(r2, list(mean = mean, sd = sd))
#> # A tibble: 1 × 3
#> r2_mean r2_sd r2_list
#> <dbl> <dbl> <list>
#> 1 0.0648 0.0488 <dbl [48]>