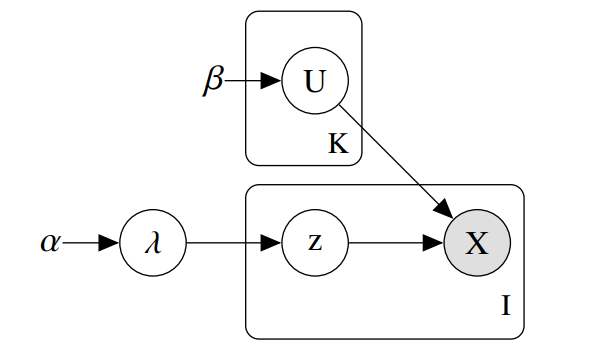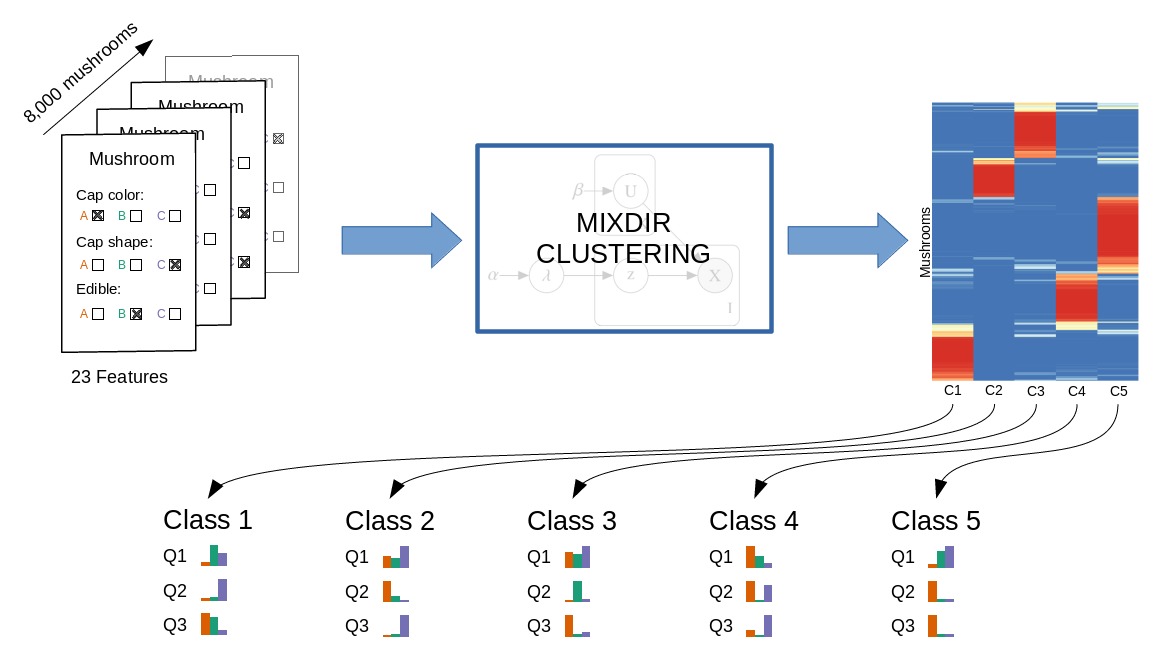
The goal of mixdir is to cluster high dimensional categorical datasets.
It can
mixdir(select_latent=TRUE))A detailed description of the algorithm and the features of the package can be found in the the accompanying paper. If you find the package useful please cite
C. Ahlmann-Eltze and C. Yau, “MixDir: Scalable Bayesian Clustering for High-Dimensional Categorical Data”, 2018 IEEE 5th International Conference on Data Science and Advanced Analytics (DSAA), Turin, Italy, 2018, pp. 526-539.
install.packages("mixdir")
# Or to get the latest version from github
devtools::install_github("const-ae/mixdir")Clustering the mushroom data set.

# Loading the library and the data
library(mixdir)
set.seed(1)
data("mushroom")
# High dimensional dataset: 8124 mushroom and 23 different features
mushroom[1:10, 1:5]
#> bruises cap-color cap-shape cap-surface edible
#> 1 bruises brown convex smooth poisonous
#> 2 bruises yellow convex smooth edible
#> 3 bruises white bell smooth edible
#> 4 bruises white convex scaly poisonous
#> 5 no gray convex smooth edible
#> 6 bruises yellow convex scaly edible
#> 7 bruises white bell smooth edible
#> 8 bruises white bell scaly edible
#> 9 bruises white convex scaly poisonous
#> 10 bruises yellow bell smooth edibleCalling the clustering function mixdir on a subset of
the data:
# Clustering into 3 latent classes
result <- mixdir(mushroom[1:1000, 1:5], n_latent=3)Analyzing the result
# Latent class of of first 10 mushrooms
head(result$pred_class, n=10)
#> [1] 3 1 1 3 2 1 1 1 3 1
# Soft Clustering for first 10 mushrooms
head(result$class_prob, n=10)
#> [,1] [,2] [,3]
#> [1,] 3.103495e-07 1.055098e-05 9.999891e-01
#> [2,] 9.998594e-01 4.683764e-06 1.359291e-04
#> [3,] 9.998944e-01 3.111462e-06 1.025194e-04
#> [4,] 5.778033e-04 7.114603e-08 9.994221e-01
#> [5,] 3.662625e-07 9.999992e-01 4.183025e-07
#> [6,] 9.996461e-01 8.764031e-08 3.537838e-04
#> [7,] 9.998944e-01 3.111462e-06 1.025194e-04
#> [8,] 9.997331e-01 5.822320e-08 2.668420e-04
#> [9,] 5.778033e-04 7.114603e-08 9.994221e-01
#> [10,] 9.999999e-01 5.850067e-09 9.845112e-08
pheatmap::pheatmap(result$class_prob, cluster_cols=FALSE,
labels_col = paste("Class", 1:3))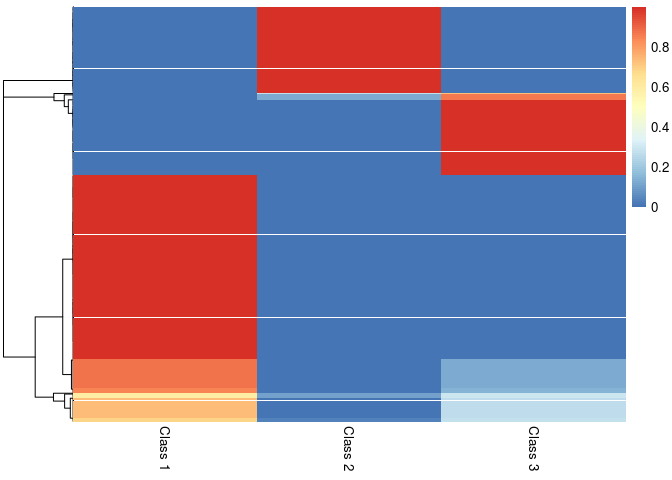
# Structure of latent class 1
# (bruises, cap color either yellow or white, edible etc.)
purrr::map(result$category_prob, 1)
#> $bruises
#> bruises no
#> 0.9998223256 0.0001776744
#>
#> $`cap-color`
#> brown gray red white yellow
#> 0.0001775934 0.0001819672 0.0001776373 0.4079822666 0.5914805356
#>
#> $`cap-shape`
#> bell convex flat sunken
#> 0.3926736 0.4767291 0.1304197 0.0001776
#>
#> $`cap-surface`
#> fibrous scaly smooth
#> 0.0568571 0.4871396 0.4560033
#>
#> $edible
#> edible poisonous
#> 0.9998223174 0.0001776826
# The most predicitive features for each class
find_predictive_features(result, top_n=3)
#> column answer class probability
#> 19 cap-color yellow 1 0.9993990
#> 22 cap-shape bell 1 0.9990947
#> 1 bruises bruises 1 0.7089533
#> 48 edible poisonous 3 0.9980468
#> 15 cap-color red 3 0.8462032
#> 9 cap-color brown 3 0.6473043
#> 5 bruises no 2 0.9990364
#> 11 cap-color gray 2 0.9978218
#> 32 cap-shape sunken 2 0.9936162
# For example: if all I know about a mushroom is that it has a
# yellow cap, then I am 99% certain that it will be in class 1
predict(result, c(`cap-color`="yellow"))
#> [,1] [,2] [,3]
#> [1,] 0.999399 0.0003004692 0.0003004907
# Note the most predictive features are different from the most typical ones
find_typical_features(result, top_n=3)
#> column answer class probability
#> 1 bruises bruises 1 0.9998223
#> 43 edible edible 1 0.9998223
#> 19 cap-color yellow 1 0.5914805
#> 3 bruises bruises 3 0.9995546
#> 27 cap-shape convex 3 0.7460615
#> 9 cap-color brown 3 0.6746224
#> 44 edible edible 2 0.9995310
#> 5 bruises no 2 0.9713177
#> 35 cap-surface fibrous 2 0.7355413Dimensionality Reduction
# Defining Features
def_feat <- find_defining_features(result, mushroom[1:1000, 1:5], n_features = 3)
print(def_feat)
#> $features
#> [1] "cap-color" "bruises" "edible"
#>
#> $quality
#> [1] 74.35146
# Plotting the most important features gives an immediate impression
# how the cluster differ
plot_features(def_feat$features, result$category_prob)
#> Loading required namespace: ggplot2
#> Loading required namespace: tidyr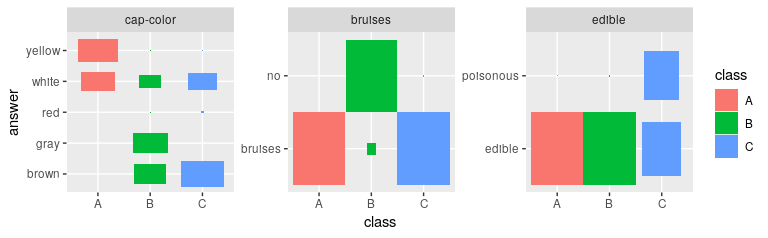
The package implements a variational inference algorithm to solve a Bayesian latent class model (LCM).