
Julia (mixed-effects) regression modelling from R. Powered by the {JuliaConnectoR}
R package and Julia libraries GLM, StatsModels, and
MixedModels.
You can install the development version of {jlme} from
GitHub with:
# install.packages("remotes")
remotes::install_github("yjunechoe/jlme"){jlme} is experimental and under active development: see
NEWS.md
for the latest updates.
library(jlme)
jlme_setup()Using {jlme} requires a prior installation of the
Julia programming language, which can be downloaded
from either the official website or
using the command line utility juliaup.
If you are encountering issues with setting up Julia, please make
sure that you’re have the latest version (>=1.1.4) of the {JuliaConnectoR}
package installed and see ?JuliaConnectoR::`Julia-Setup`
for troubleshooting.
Once set up, (g)lm() and (g)lmer
complements in Julia are available via jlm() and
jlmer(), respectively.
jlm() with lm()/glm()
syntax:
# lm(mpg ~ hp, mtcars)
jlm(mpg ~ hp, mtcars)
#> <Julia object of type StatsModels.TableRegressionModel>
#>
#> mpg ~ 1 + hp
#>
#> ────────────────────────────────────────────────────────────────────────────
#> Coef. Std. Error z Pr(>|z|) Lower 95% Upper 95%
#> ────────────────────────────────────────────────────────────────────────────
#> (Intercept) 30.0989 1.63392 18.42 <1e-75 26.8964 33.3013
#> hp -0.0682283 0.0101193 -6.74 <1e-10 -0.0880617 -0.0483948
#> ────────────────────────────────────────────────────────────────────────────Contrasts in factor columns are preserved:
x <- mtcars
# Sum code `am`
x$am_sum <- factor(x$am)
contrasts(x$am_sum) <- contr.sum(2)
# Helmert code `cyl`
x$cyl_helm <- factor(x$cyl)
contrasts(x$cyl_helm) <- contr.helmert(3)
colnames(contrasts(x$cyl_helm)) <- c("4vs6", "4&6vs8")
jlm(mpg ~ am_sum + cyl_helm, x)
#> <Julia object of type StatsModels.TableRegressionModel>
#>
#> mpg ~ 1 + am_sum + cyl_helm
#>
#> ───────────────────────────────────────────────────────────────────────────────
#> Coef. Std. Error z Pr(>|z|) Lower 95% Upper 95%
#> ───────────────────────────────────────────────────────────────────────────────
#> (Intercept) 20.6739 0.572633 36.10 <1e-99 19.5516 21.7963
#> am_sum: 1 -1.27998 0.648789 -1.97 0.0485 -2.55158 -0.00837293
#> cyl_helm: 4vs6 -3.07806 0.767861 -4.01 <1e-04 -4.58304 -1.57308
#> cyl_helm: 4&6vs8 -2.32983 0.414392 -5.62 <1e-07 -3.14203 -1.51764
#> ───────────────────────────────────────────────────────────────────────────────jlmer() with lmer()/glmer()
syntax:
# lme4::lmer(Reaction ~ Days + (Days | Subject), lme4::sleepstudy)
jlmer(Reaction ~ Days + (Days | Subject), lme4::sleepstudy, REML = TRUE)
#> <Julia object of type LinearMixedModel>
#>
#> Linear mixed model fit by REML
#> Reaction ~ 1 + Days + (1 + Days | Subject)
#> REML criterion at convergence: 1743.6282719599653
#>
#> Variance components:
#> Column Variance Std.Dev. Corr.
#> Subject (Intercept) 612.10016 24.74066
#> Days 35.07171 5.92214 +0.07
#> Residual 654.94001 25.59180
#> Number of obs: 180; levels of grouping factors: 18
#>
#> Fixed-effects parameters:
#> ──────────────────────────────────────────────────
#> Coef. Std. Error z Pr(>|z|)
#> ──────────────────────────────────────────────────
#> (Intercept) 251.405 6.8246 36.84 <1e-99
#> Days 10.4673 1.54579 6.77 <1e-10
#> ──────────────────────────────────────────────────# lme4::glmer(r2 ~ Anger + Gender + (1 | id), lme4::VerbAgg, family = "binomial")
jlmer(r2 ~ Anger + Gender + (1 | id), lme4::VerbAgg, family = "binomial")
#> <Julia object of type GeneralizedLinearMixedModel>
#>
#> Generalized Linear Mixed Model fit by maximum likelihood (nAGQ = 1)
#> r2 ~ 1 + Anger + Gender + (1 | id)
#> Distribution: Bernoulli{Float64}
#> Link: LogitLink()
#>
#> logLik deviance AIC AICc BIC
#> -4748.2525 9496.5050 9504.5050 9504.5102 9532.2401
#>
#> Variance components:
#> Column VarianceStd.Dev.
#> id (Intercept) 1.12074 1.05865
#>
#> Number of obs: 7584; levels of grouping factors: 316
#>
#> Fixed-effects parameters:
#> ────────────────────────────────────────────────────
#> Coef. Std. Error z Pr(>|z|)
#> ────────────────────────────────────────────────────
#> (Intercept) -1.10115 0.280681 -3.92 <1e-04
#> Anger 0.0462741 0.0134906 3.43 0.0006
#> Gender: M 0.260057 0.153847 1.69 0.0910
#> ────────────────────────────────────────────────────Supports {broom}-style tidy() and
glance() methods for Julia regression models.
Get information about model components with tidy()
# Note that MixedModels/`jlmer()` defaults to ML (REML=false)
jmod <- jlmer(Reaction ~ Days + (Days | Subject), lme4::sleepstudy)
tidy(jmod)
#> effect group term estimate std.error statistic
#> 1 fixed <NA> (Intercept) 251.40510485 6.632258 37.906414
#> 2 fixed <NA> Days 10.46728596 1.502236 6.967806
#> 12 ran_pars Subject sd__(Intercept) 23.78046792 NA NA
#> 3 ran_pars Subject cor__(Intercept).Days 0.08133207 NA NA
#> 21 ran_pars Subject sd__Days 5.71682816 NA NA
#> 11 ran_pars Residual sd__Observation 25.59182388 NA NA
#> p.value
#> 1 2.017794e-314
#> 2 3.219214e-12
#> 12 NA
#> 3 NA
#> 21 NA
#> 11 NAGet goodness-of-fit measures of a model with
glance()
glance(jmod)
#> nobs df sigma logLik AIC BIC deviance df.residual
#> 1 180 6 25.59182 -875.9697 1763.939 1783.097 1751.939 174Check singular fit
issingular(jmod)
#> [1] FALSEList all properties of a MixedModel object (properties are accessible
via $)
propertynames(jmod)
#> [1] "A" "b" "beta" "betas" "corr" "dims"
#> [7] "feterm" "formula" "L" "lambda" "lowerbd" "objective"
#> [13] "optsum" "parmap" "PCA" "pvalues" "rePCA" "reterms"
#> [19] "sigma" "sigmarhos" "sigmas" "sqrtwts" "stderror" "theta"
#> [25] "u" "vcov" "X" "Xymat" "y" "β"
#> [31] "βs" "θ" "λ" "σ" "σs" "σρs"Check optimization summary
jmod$optsum
#> <Julia object of type OptSummary{Float64}>
#> Initial parameter vector: [1.0, 0.0, 1.0]
#> Initial objective value: 1784.642296192471
#>
#> Optimizer (from NLopt): LN_BOBYQA
#> Lower bounds: [0.0, -Inf, 0.0]
#> ftol_rel: 1.0e-12
#> ftol_abs: 1.0e-8
#> xtol_rel: 0.0
#> xtol_abs: [1.0e-10, 1.0e-10, 1.0e-10]
#> initial_step: [0.75, 1.0, 0.75]
#> maxfeval: -1
#> maxtime: -1.0
#>
#> Function evaluations: 57
#> Final parameter vector: [0.9292213025841999, 0.018168360086059557, 0.22264488361408383]
#> Final objective value: 1751.9393444646894
#> Return code: FTOL_REACHEDFunctions parametricbootstrap() and
profilelikelihood() can be used to assess the variability
of parameter estimates.
Experimental support for MixedModels.parametricbootstrap
via parametricbootstrap():
samp <- parametricbootstrap(jmod, nsim = 100L, seed = 42L)
samp
#> <Julia object of type MixedModelBootstrap{Float64}>
#> MixedModelBootstrap with 100 samples
#> parameter min q25 median mean q75 max
#> ┌────────────────────────────────────────────────────────────────────────────
#> 1 │ β1 228.0 246.58 250.863 250.977 256.082 264.406
#> 2 │ β2 6.72055 9.91274 10.8382 10.696 11.6725 13.7769
#> 3 │ σ 21.6856 24.6536 25.6211 25.838 26.7439 30.5968
#> 4 │ σ1 3.57556 17.8908 22.1421 21.5799 25.6259 31.9446
#> 5 │ σ2 1.63637 4.53406 5.36349 5.47536 6.38651 8.34052
#> 6 │ ρ1 -0.739267 -0.226513 0.120712 0.107686 0.389495 1.0
#> 7 │ θ1 0.146373 0.675859 0.845334 0.839596 1.02343 1.29287
#> 8 │ θ2 -0.18147 -0.0508505 0.0248657 0.0155747 0.0708033 0.226194
#> 9 │ θ3 0.0 0.156785 0.193042 0.188678 0.238505 0.334108tidy(samp)
#> effect group term estimate conf.low conf.high
#> 1 fixed <NA> (Intercept) 251.40510485 240.8972974 263.280668
#> 2 fixed <NA> Days 10.46728596 7.7302901 13.540729
#> 5 ran_pars Subject sd__(Intercept) 23.78046792 12.5974975 31.944579
#> 4 ran_pars Subject cor__(Intercept).Days 0.08133207 -0.5948863 1.000000
#> 6 ran_pars Subject sd__Days 5.71682816 3.6653412 8.073549
#> 3 ran_pars Residual sd__Observation 25.59182388 22.4968576 29.124647Experimental support for MixedModels.profile
via profilelikelihood():
prof <- profilelikelihood(jmod)
prof
#> <Julia object of type MixedModelProfile{Float64}>
#> MixedModelProfile -- Table with 11 columns and 176 rows:
#> p ζ β1 β2 σ σ1 σ2 ρ1 ⋯
#> ┌──────────────────────────────────────────────────────────────────────────
#> 1 │ σ -4.365 251.405 10.4673 20.1933 25.5128 5.97319 -0.0159232 ⋯
#> 2 │ σ -3.77902 251.405 10.4673 20.8002 25.3434 5.94786 -0.00711109 ⋯
#> 3 │ σ -3.20526 251.405 10.4673 21.4255 25.1627 5.92133 0.00263598 ⋯
#> 4 │ σ -2.64336 251.405 10.4673 22.0695 24.9702 5.89214 0.0129078 ⋯
#> 5 │ σ -2.09298 251.405 10.4673 22.7328 24.7636 5.86167 0.0241968 ⋯
#> 6 │ σ -1.55378 251.405 10.4673 23.4162 24.5426 5.82871 0.0366992 ⋯
#> 7 │ σ -1.02542 251.405 10.4673 24.12 24.3063 5.79389 0.0501628 ⋯
#> 8 │ σ -0.507597 251.405 10.4673 24.845 24.0525 5.75669 0.0649969 ⋯
#> 9 │ σ 0.0 251.405 10.4673 25.5918 23.7805 5.71683 0.0813321 ⋯
#> 10 │ σ 0.497684 251.405 10.4673 26.3611 23.4885 5.6743 0.0993046 ⋯
#> 11 │ σ 0.985728 251.405 10.4673 27.1534 23.1743 5.62882 0.119204 ⋯
#> 12 │ σ 1.46442 251.405 10.4673 27.9696 22.836 5.5802 0.141262 ⋯
#> 13 │ σ 1.93402 251.405 10.4673 28.8104 22.4727 5.52809 0.165869 ⋯
#> 14 │ σ 2.39481 251.405 10.4673 29.6763 22.0799 5.47226 0.193415 ⋯
#> 15 │ σ 2.84704 251.405 10.4673 30.5684 21.6557 5.41243 0.224387 ⋯
#> 16 │ σ 3.29094 251.405 10.4673 31.4872 21.196 5.34818 0.259442 ⋯
#> 17 │ σ 3.72677 251.405 10.4673 32.4337 20.6956 5.27882 0.299408 ⋯
#> ⋮ │ ⋮ ⋮ ⋮ ⋮ ⋮ ⋮ ⋮ ⋮ ⋱tidy(prof)
#> effect group term estimate conf.low conf.high
#> 1 fixed <NA> (Intercept) 251.405105 237.680694 265.129516
#> 2 fixed <NA> Days 10.467286 7.358653 13.575919
#> 12 ran_pars Subject sd__(Intercept) 23.780468 14.381431 37.718099
#> 21 ran_pars Subject sd__Days 5.716828 0.000000 8.753389
#> 11 ran_pars Residual sd__Observation 25.591824 22.898262 28.858001Functions jl_get() and jl_put() transfers
data between R and Julia.
Example 1: extract PCA of random effects and return as an R list:
jmod$rePCA
#> <Julia object of type @NamedTuple{Subject::Vector{Float64}}>
#> (Subject = [0.5406660352881864, 1.0],)jl_get(jmod$rePCA)
#> $Subject
#> [1] 0.540666 1.000000Example 2: extract fitlog and plot
fitlog <- jl_get(jl("refit!(deepcopy(x); thin=1)", x = jmod)$optsum$fitlog)
thetas <- t(sapply(fitlog, `[[`, 1))
matplot(thetas, type = "o", xlab = "iterations")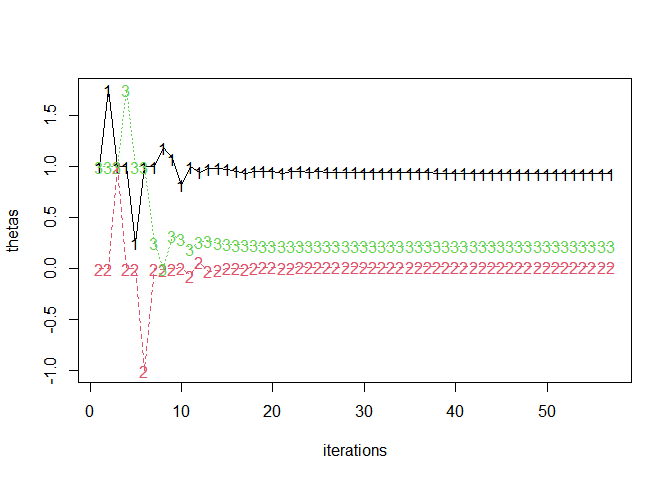
See information about the running Julia environment (e.g., the list
of loaded Julia libraries) with jlme_status():
jlme_status()
#> jlme 0.4.1
#> R version 4.4.1 (2024-06-14 ucrt)
#> Julia Version 1.10.5
#> Commit 6f3fdf7b36 (2024-08-27 14:19 UTC)
#> Build Info:
#> Official https://julialang.org/ release
#> Platform Info:
#> OS: Windows (x86_64-w64-mingw32)
#> CPU: 8 × 11th Gen Intel(R) Core(TM) i7-1165G7 @ 2.80GHz
#> WORD_SIZE: 64
#> LIBM: libopenlibm
#> LLVM: libLLVM-15.0.7 (ORCJIT, tigerlake)
#> Threads: 1 default, 0 interactive, 1 GC (on 8 virtual cores)
#> Status `C:\Users\jchoe\AppData\Local\Temp\jl_NI67vu\Project.toml`
#> [38e38edf] GLM v1.9.0
#> [ff71e718] MixedModels v4.27.0
#> [3eaba693] StatsModels v0.7.4
#> [9a3f8284] RandomOn setup, {jlme} loads GLM, StatsModels, and
MixedModels,
as well as those specified in jlme_setup(add). Other
libraries such as Random (required for
parametricbootstrap()) are loaded on an as-needed
basis.
{JuliaConnectoR}While users will typically not need to interact with
{JuliaConnectoR} directly, it may be useful for extending
{jlme} features with other packages in the Julia modelling
ecosystem. A simple way to do that is to use juliaImport(),
which creates makeshift bindings to any Julia library.
Here’s an example replicating a workflow using Effects.empairs
for post-hoc pairwise comparisons:
# New model: 2 (M/F) by 3 (curse/scold/shout) factorial
jmod2 <- jlmer(
r2 ~ Gender * btype + (1 | id),
data = lme4::VerbAgg,
family = "binomial"
)
jmod2
#> <Julia object of type GeneralizedLinearMixedModel>
#>
#> Generalized Linear Mixed Model fit by maximum likelihood (nAGQ = 1)
#> r2 ~ 1 + Gender + btype + Gender & btype + (1 | id)
#> Distribution: Bernoulli{Float64}
#> Link: LogitLink()
#>
#> logLik deviance AIC AICc BIC
#> -4341.4933 8682.9867 8696.9867 8697.0014 8745.5232
#>
#> Variance components:
#> Column VarianceStd.Dev.
#> id (Intercept) 1.52160 1.23353
#>
#> Number of obs: 7584; levels of grouping factors: 316
#>
#> Fixed-effects parameters:
#> ─────────────────────────────────────────────────────────────────
#> Coef. Std. Error z Pr(>|z|)
#> ─────────────────────────────────────────────────────────────────
#> (Intercept) 0.738396 0.0956957 7.72 <1e-13
#> Gender: M 0.404282 0.201612 2.01 0.0449
#> btype: scold -1.01214 0.0742669 -13.63 <1e-41
#> btype: shout -1.77376 0.0782721 -22.66 <1e-99
#> Gender: M & btype: scold 0.130832 0.156315 0.84 0.4026
#> Gender: M & btype: shout -0.533658 0.168832 -3.16 0.0016
#> ─────────────────────────────────────────────────────────────────library(JuliaConnectoR)
# First call downloads the library (takes a minute)
Effects <- juliaImport("Effects")
# Call `Effects.empairs` using R syntax `Effects$empairs()`
pairwise <- Effects$empairs(jmod2, dof = glance(jmod2)$df.residual)
pairwise
#> <Julia object of type DataFrames.DataFrame>
#> 15×7 DataFrame
#> Row │ Gender btype r2: Y err dof t Pr(>|t|) ⋯
#> │ String String Float64 Float64 Int64 Float64 Float64 ⋯
#> ─────┼──────────────────────────────────────────────────────────────────────────
#> 1 │ F > M curse -0.404282 0.201612 7577 -2.00524 0.0449725 ⋯
#> 2 │ F curse > scold 1.01214 0.13461 7577 7.51901 6.15243e-
#> 3 │ F > M curse > scold 0.477022 0.196994 7577 2.4215 0.0154799
#> 4 │ F curse > shout 1.77376 0.136284 7577 13.0152 2.57693e-
#> 5 │ F > M curse > shout 1.90314 0.202352 7577 9.4051 6.75459e- ⋯
#> 6 │ M > F curse > scold 1.41642 0.201127 7577 7.0424 2.05522e-
#> 7 │ M curse > scold 0.881304 0.247263 7577 3.56424 0.0003671
#> 8 │ M > F curse > shout 2.17805 0.202251 7577 10.769 7.53779e-
#> 9 │ M curse > shout 2.30742 0.251552 7577 9.17273 5.84715e- ⋯
#> 10 │ F > M scold -0.535114 0.196498 7577 -2.72326 0.0064789
#> 11 │ F scold > shout 0.761627 0.135565 7577 5.61817 1.99834e-
#> 12 │ F > M scold > shout 0.891002 0.201868 7577 4.41378 1.02988e-
#> 13 │ M > F scold > shout 1.29674 0.197648 7577 6.56086 5.70179e- ⋯
#> 14 │ M scold > shout 1.42612 0.247866 7577 5.75357 9.07797e-
#> 15 │ F > M shout 0.129376 0.202988 7577 0.637355 0.523913
#> 1 column omittedNote that Julia DataFrame objects such as the one above
can be collected into an R data frame using
as.data.frame(). This lets you, for example, apply p-value
corrections using the familiar p.adjust() function in R,
though the option
to do that exists in Julia as well.
pairwise_df <- as.data.frame(pairwise)
cbind(
pairwise_df[, 1:2],
round(pairwise_df[, 3:4], 2),
pvalue = format.pval(p.adjust(pairwise_df[, 7], "bonferroni"), 1)
)
#> Gender btype r2..Y err pvalue
#> 1 F > M curse -0.40 0.20 0.675
#> 2 F curse > scold 1.01 0.13 9e-13
#> 3 F > M curse > scold 0.48 0.20 0.232
#> 4 F curse > shout 1.77 0.14 <2e-16
#> 5 F > M curse > shout 1.90 0.20 <2e-16
#> 6 M > F curse > scold 1.42 0.20 3e-11
#> 7 M curse > scold 0.88 0.25 0.006
#> 8 M > F curse > shout 2.18 0.20 <2e-16
#> 9 M curse > shout 2.31 0.25 <2e-16
#> 10 F > M scold -0.54 0.20 0.097
#> 11 F scold > shout 0.76 0.14 3e-07
#> 12 F > M scold > shout 0.89 0.20 2e-04
#> 13 M > F scold > shout 1.30 0.20 9e-10
#> 14 M scold > shout 1.43 0.25 1e-07
#> 15 F > M shout 0.13 0.20 1.000MixedModels.jl supports various display
formats for mixed-effects models which are available in
{jlme} via the format argument of
print():
# Rendered via {knitr} with chunk option `results="asis"`
print(jmod, format = "markdown")| Est. | SE | z | p | σ_Subject | |
|---|---|---|---|---|---|
| (Intercept) | 251.4051 | 6.6323 | 37.91 | <1e-99 | 23.7805 |
| Days | 10.4673 | 1.5022 | 6.97 | <1e-11 | 5.7168 |
| Residual | 25.5918 |
Be sure to pass integers (vs. doubles) to Julia functions that expect Integer type:
jl_put(1)
#> <Julia object of type Float64>
#> 1.0jl_put(1L)
#> <Julia object of type Int64>
#> 1The Dict (dictionary) data type is common and relevant
for modelling workflows in Julia. {jlme} offers
jl_dict() as an opinionated constructor for that
usecase:
# Note use of `I()` to protect length-1 values as scalar
jl_dict(a = 1:2, b = "three", c = I(4.5))
#> <Julia object of type Dict{Symbol, Any}>
#> Dict{Symbol, Any} with 3 entries:
#> :a => [1, 2]
#> :b => ["three"]
#> :c => 4.5See ?JuliaConnectoR::`JuliaConnectoR-package` for a
comprehensive list of data type conversion rules.
Using MKL.jl
or AppleAccelerate.jl
may improve model fitting performance (but see the system requirements
first). This should be supplied to the add argument to
jlme_setup(), to ensure that they are loaded first, prior
to other packages.
# Not run
jlme_setup(add = "MKL", restart = TRUE)
jlme_status() # Should now see MKL loaded hereIf the data is large, a sizable overhead will come from transferring
the data from R to Julia. If you are also looking to fit many models to
the same data, you should first filter to keep only the columns you need
and then use jl_data() to send the data to Julia. The Julia
object can then be used in place of an R data frame.
data_r <- mtcars
# Keep only columns you need + convert with `jl_data()`
data_julia <- jl_data(data_r[, c("mpg", "am")])
jlm(mpg ~ am, data_julia)
#> <Julia object of type StatsModels.TableRegressionModel>
#>
#> mpg ~ 1 + am
#>
#> ────────────────────────────────────────────────────────────────────────
#> Coef. Std. Error z Pr(>|z|) Lower 95% Upper 95%
#> ────────────────────────────────────────────────────────────────────────
#> (Intercept) 17.1474 1.1246 15.25 <1e-51 14.9432 19.3515
#> am 7.24494 1.76442 4.11 <1e-04 3.78674 10.7031
#> ────────────────────────────────────────────────────────────────────────If your data has custom contrasts, you can use
jl_contrasts() to also convert that to Julia first before
passing it to the model.
data_r$am <- as.factor(data_r$am)
contrasts(data_r$am) <- contr.sum(2)
data_julia <- jl_data(data_r[, c("mpg", "am")])
contrasts_julia <- jl_contrasts(data_r)
jlm(mpg ~ am, data_julia, contrasts = contrasts_julia)
#> <Julia object of type StatsModels.TableRegressionModel>
#>
#> mpg ~ 1 + am
#>
#> ────────────────────────────────────────────────────────────────────────
#> Coef. Std. Error z Pr(>|z|) Lower 95% Upper 95%
#> ────────────────────────────────────────────────────────────────────────
#> (Intercept) 20.7698 0.882211 23.54 <1e-99 19.0407 22.4989
#> am: 1 -3.62247 0.882211 -4.11 <1e-04 -5.35157 -1.89337
#> ────────────────────────────────────────────────────────────────────────If you spend non-negligible time fitting regression models for your work, please just learn Julia! It’s a great high-level language that feels close to R in syntax and its REPL-based workflow.
The JuliaConnectoR R package for powering the R interface to Julia.
The Julia packages GLM.jl, StatsModels, and MixedModels.jl for interfaces to and implementations of (mixed effects) regression models.
Other R interfaces to MixedModels. It has come to my attention
after working on this for a while and publishing it to CRAN that there
were prior efforts by Mika
Braginsky in {jglmm} and by
Philip Alday in {jlme}. These
packages/scripts predate {JuliaConnectoR} (which I find to
be leaner and more robust) and are instead built on top of {JuliaCall}.