The intensitynet package provides tools to analyze point patterns in
space over planar network structures derived from graph-related
intensity measures for undirected, directed, and mixed networks. This
package is based on the research done by Eckardt, M., Mateu, J.
presented in the following papers:
Eckardt, M., Mateu, J.
Point Patterns Occurring on Complex Structures in Space and Space-Time:
An Alternative Network Approach. Journal of Computational and Graphical
Statistics 27. 312-322 (2017).
10.1080/10618600.2017.1391695
Eckardt, M., Mateu, J. Second-order and local characteristics
of network intensity functions. TEST 30, 318-340 (2021).
10.1007/s11749-020-00720-4
You can install the released version of intensitynet from CRAN with:
install.packages("intensitynet")And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("LlagosteraPol/intensitynet")This is an example that shows you how to set up intensitynet and calculate and plot the Geary-c correlation:
library(intensitynet)
library(spatstat)
#> Loading required package: spatstat.data
#> Loading required package: spatstat.geom
#> spatstat.geom 2.3-1
#> Loading required package: spatstat.core
#> Loading required package: nlme
#> Loading required package: rpart
#> spatstat.core 2.3-2
#> Loading required package: spatstat.linnet
#> spatstat.linnet 2.3-1
#>
#> spatstat 2.3-0 (nickname: 'That's not important right now')
#> For an introduction to spatstat, type 'beginner'
data(chicago)
chicago_df <- as.data.frame(chicago[["data"]]) # Get as dataframe the data from Chicago
# Get the adjacency matrix. One way is to create an igraph object from the edge coordinates.
edges <- cbind(chicago[["domain"]][["from"]], chicago[["domain"]][["to"]])
chicago_net <- igraph::graph_from_edgelist(edges)
# And then use the igraph function 'as_adjacency_matrix'
chicago_adj_mtx <- as.matrix(igraph::as_adjacency_matrix(chicago_net))
chicago_node_coords <- data.frame(xcoord = chicago[["domain"]][["vertices"]][["x"]],
ycoord = chicago[["domain"]][["vertices"]][["y"]])
# Create the intensitynet object, in this case will be undirected
intnet_chicago <- intensitynet(chicago_adj_mtx,
node_coords = chicago_node_coords,
event_data = chicago_df)
intnet_chicago <- RelateEventsToNetwork(intnet_chicago)
#> Calculating edge intensities...
#> ================================================================================
#> Calculating node intensities...
#> ================================================================================
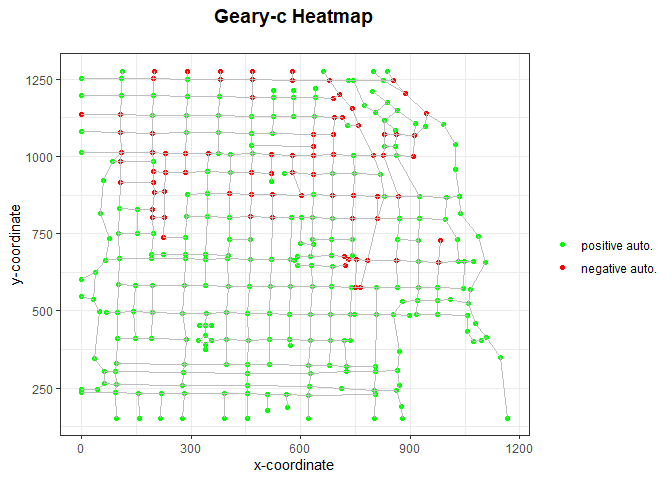
data_geary <- NodeLocalCorrelation(intnet_chicago, dep_type = 'geary', intensity = igraph::vertex_attr(intnet_chicago$graph)$intensity)
geary_c <- data_geary$correlation
intnet_chicago <- data_geary$intnet
PlotHeatmap(intnet_chicago, heattype='geary')