
This is an R wrapper around the COVID Tracking Project API. It provides updates on the spread of the virus in the US with a few simple functions.
install.packages("covid19us")Or the dev version:
devtools::install_github("aedobbyn/covid19us")library(covid19us)Get the most recent COVID-19 top-line data for the country:
get_us_current()
#> # A tibble: 1 x 18
#> positive negative pending hospitalized_cu… hospitalized_cu… in_icu_currently
#> <int> <int> <int> <int> <int> <int>
#> 1 1520778 10713209 2944 41174 159634 9829
#> # … with 12 more variables: in_icu_cumulative <int>,
#> # on_ventilator_currently <int>, on_ventilator_cumulative <int>,
#> # recovered <int>, hash <chr>, last_modified <chr>, death <int>,
#> # hospitalized <int>, total <int>, total_test_results <int>, notes <chr>,
#> # request_datetime <dttm>Or the same by state:
get_states_current()
#> # A tibble: 56 x 30
#> state positive positive_score negative_score negative_regula…
#> <chr> <int> <int> <int> <int>
#> 1 AK 399 1 1 1
#> 2 AL 12376 1 1 0
#> 3 AR 4923 1 1 1
#> 4 AZ 14566 1 1 0
#> 5 CA 81795 1 1 0
#> 6 CO 22202 1 1 1
#> 7 CT 38430 1 1 1
#> 8 DC 7434 1 1 1
#> 9 DE 8037 1 1 1
#> 10 FL 46944 1 1 1
#> # … with 46 more rows, and 25 more variables: commercial_score <int>,
#> # grade <chr>, score <int>, notes <chr>, data_quality_grade <chr>,
#> # negative <int>, pending <int>, hospitalized_currently <int>,
#> # hospitalized_cumulative <int>, in_icu_currently <int>,
#> # in_icu_cumulative <int>, on_ventilator_currently <int>,
#> # on_ventilator_cumulative <int>, recovered <int>, last_update <dttm>,
#> # check_time <dttm>, death <int>, hospitalized <int>, total <int>,
#> # total_test_results <int>, fips <chr>, date_modified <dttm>,
#> # date_checked <dttm>, hash <chr>, request_datetime <dttm>Daily state counts can be filtered by state and/or date:
get_states_daily(
state = "NY",
date = "2020-03-17"
)
#> # A tibble: 1 x 27
#> date state positive negative pending hospitalized_cu… hospitalized_cu…
#> <date> <chr> <int> <int> <int> <int> <int>
#> 1 2020-03-17 NY 1700 5506 NA 325 NA
#> # … with 20 more variables: in_icu_currently <int>, in_icu_cumulative <int>,
#> # on_ventilator_currently <int>, on_ventilator_cumulative <int>,
#> # recovered <int>, data_quality_grade <chr>, last_update <dttm>, hash <chr>,
#> # date_checked <dttm>, death <int>, hospitalized <int>, total <int>,
#> # total_test_results <int>, fips <chr>, death_increase <int>,
#> # hospitalized_increase <int>, negative_increase <int>,
#> # positive_increase <int>, total_test_results_increase <int>,
#> # request_datetime <dttm>For data in long format:
(dat <- refresh_covid19us())
#> # A tibble: 80,123 x 7
#> date location location_type location_code location_code_t… data_type
#> <date> <chr> <chr> <chr> <chr> <chr>
#> 1 2020-05-19 AK state 02 fips_code positive
#> 2 2020-05-19 AK state 02 fips_code negative
#> 3 2020-05-19 AK state 02 fips_code pending
#> 4 2020-05-19 AK state 02 fips_code hospital…
#> 5 2020-05-19 AK state 02 fips_code hospital…
#> 6 2020-05-19 AK state 02 fips_code in_icu_c…
#> 7 2020-05-19 AK state 02 fips_code in_icu_c…
#> 8 2020-05-19 AK state 02 fips_code on_venti…
#> 9 2020-05-19 AK state 02 fips_code on_venti…
#> 10 2020-05-19 AK state 02 fips_code recovered
#> # … with 80,113 more rows, and 1 more variable: value <int>Which can be easier to plot
library(dplyr)
library(ggplot2)
dat %>%
filter(
location == "NY" &
data_type %in%
c(
"positive_increase",
"total_test_results_increase",
"death_increase",
"hospitalized_increase"
)
) %>%
mutate(
Type = data_type %>%
stringr::str_replace_all("_", " ") %>%
stringr::str_to_title()
) %>%
arrange(date) %>%
ggplot(aes(date, value, color = Type)) +
geom_smooth(se = FALSE) +
scale_x_date(date_breaks = "2 weeks") +
labs(title = "COVID in NY") +
xlab("Date") +
ylab("Value") +
theme_minimal(base_family = "Source Sans Pro")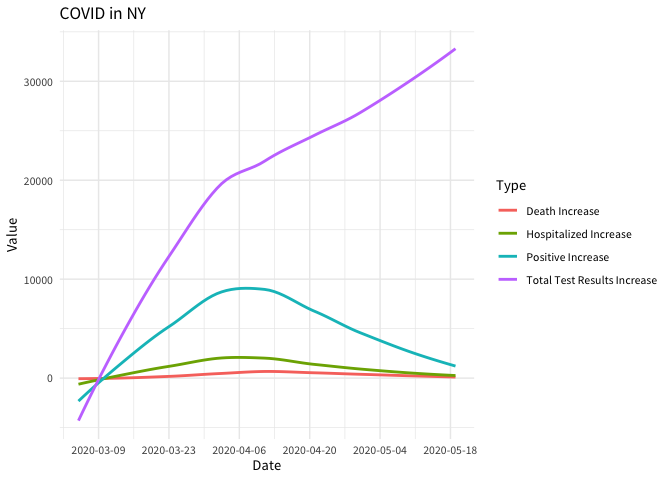
To get information about the data:
get_info_covid19us()
#> # A tibble: 1 x 10
#> data_set_name package_name function_to_get… data_details data_url license_url
#> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 covid19us covid19us refresh_covid19… Open Source… https:/… https://gi…
#> # … with 4 more variables: data_types <chr>, location_types <chr>,
#> # spatial_extent <chr>, has_geospatial_info <lgl>get_counties_info
get_info_covid19us
get_states_current
get_states_daily
get_states_info
get_tracker_urls
get_us_current
get_us_daily
refresh_covid19usAll of the data sources can be found with
get_tracker_urls()
filter column gives information about how the COVID Tracking
Project’s scraper currently scrapes data from the page (xpaths, CSS
selectors, functions used, etc.)State breakdowns include DC as well as some US territories including American Samoa (AS), Guam (GU), Northern Mariana Islands (MP), Puerto Rico (PR), and the Virgin Islands (VI)
Acronyms
Time zone used is Eastern Standard Time
PRs and bug reports / feature requests welcome. Stay safe!